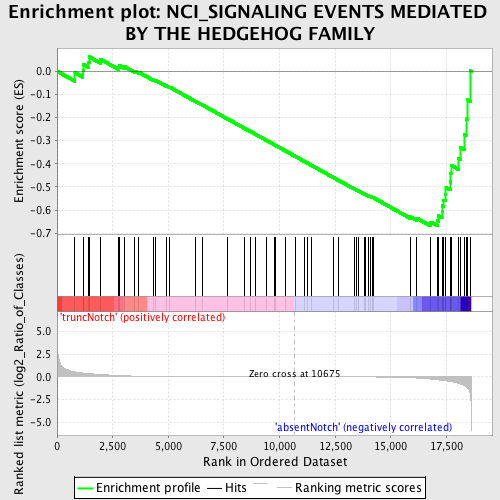
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #truncNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#truncNotch_versus_absentNotch |
| Upregulated in class | absentNotch |
| GeneSet | NCI_SIGNALING EVENTS MEDIATED BY THE HEDGEHOG FAMILY |
| Enrichment Score (ES) | -0.6686863 |
| Normalized Enrichment Score (NES) | -1.5205863 |
| Nominal p-value | 0.004149378 |
| FDR q-value | 0.32164723 |
| FWER p-Value | 0.985 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARRB2 | 2060441 | 803 | 0.543 | -0.0064 | No | ||
| 2 | ADRBK1 | 1340333 | 1163 | 0.412 | 0.0023 | No | ||
| 3 | SUFU | 6650181 | 1169 | 0.409 | 0.0298 | No | ||
| 4 | HHAT | 4200717 | 1424 | 0.348 | 0.0397 | No | ||
| 5 | PRKACA | 2640731 4050048 | 1442 | 0.342 | 0.0620 | No | ||
| 6 | LRPAP1 | 5570253 | 1957 | 0.250 | 0.0513 | No | ||
| 7 | SIN3B | 6650253 | 2753 | 0.139 | 0.0179 | No | ||
| 8 | CSNK1D | 4280280 4480167 6450600 | 2795 | 0.134 | 0.0248 | No | ||
| 9 | RBBP4 | 6650528 | 3014 | 0.113 | 0.0208 | No | ||
| 10 | SMO | 5340538 6350132 | 3483 | 0.079 | 0.0009 | No | ||
| 11 | CSNK1G1 | 840082 1230575 3940647 | 3660 | 0.069 | -0.0038 | No | ||
| 12 | PTHLH | 5290739 | 4349 | 0.046 | -0.0378 | No | ||
| 13 | GNAZ | 6130296 | 4438 | 0.044 | -0.0396 | No | ||
| 14 | PTCH1 | 430278 | 4914 | 0.034 | -0.0629 | No | ||
| 15 | GLI3 | 5690148 | 5064 | 0.031 | -0.0688 | No | ||
| 16 | GLI2 | 3060632 | 6229 | 0.017 | -0.1304 | No | ||
| 17 | LRP2 | 1400204 3290706 | 6540 | 0.014 | -0.1461 | No | ||
| 18 | HHIP | 6940438 | 7656 | 0.009 | -0.2056 | No | ||
| 19 | CDON | 540204 | 8429 | 0.006 | -0.2468 | No | ||
| 20 | PIK3R1 | 4730671 | 8693 | 0.005 | -0.2606 | No | ||
| 21 | BOC | 5270348 | 8913 | 0.005 | -0.2721 | No | ||
| 22 | MTSS1 | 780435 2370114 | 9421 | 0.003 | -0.2992 | No | ||
| 23 | FBXW11 | 6450632 | 9783 | 0.002 | -0.3185 | No | ||
| 24 | GNAI3 | 4670739 | 9818 | 0.002 | -0.3202 | No | ||
| 25 | GAS1 | 2120504 | 10284 | 0.001 | -0.3452 | No | ||
| 26 | IHH | 1980735 | 10710 | -0.000 | -0.3681 | No | ||
| 27 | XPO1 | 540707 | 11119 | -0.001 | -0.3900 | No | ||
| 28 | AKT1 | 5290746 | 11232 | -0.001 | -0.3959 | No | ||
| 29 | GLI1 | 4560176 | 11448 | -0.002 | -0.4074 | No | ||
| 30 | TGFB2 | 4920292 | 12411 | -0.005 | -0.4588 | No | ||
| 31 | PIK3CA | 6220129 | 12641 | -0.006 | -0.4708 | No | ||
| 32 | GNAI1 | 4560390 | 13372 | -0.010 | -0.5094 | No | ||
| 33 | DHH | 2120433 | 13472 | -0.010 | -0.5141 | No | ||
| 34 | SHH | 5570400 | 13531 | -0.011 | -0.5165 | No | ||
| 35 | SAP18 | 3610092 | 13801 | -0.014 | -0.5300 | No | ||
| 36 | PTCH2 | 2900092 | 13867 | -0.014 | -0.5326 | No | ||
| 37 | FOXA2 | 540338 5860441 | 14017 | -0.016 | -0.5395 | No | ||
| 38 | CSNK1A1 | 2340427 | 14088 | -0.017 | -0.5421 | No | ||
| 39 | RAB23 | 2970075 | 14193 | -0.019 | -0.5464 | No | ||
| 40 | GNAO1 | 2510292 6590487 | 14218 | -0.019 | -0.5464 | No | ||
| 41 | MAP2K1 | 840739 | 15876 | -0.103 | -0.6287 | No | ||
| 42 | GNG2 | 2230390 | 16167 | -0.135 | -0.6352 | No | ||
| 43 | CSNK1G3 | 110450 | 16790 | -0.251 | -0.6516 | Yes | ||
| 44 | RBBP7 | 430113 450450 2370309 | 17096 | -0.331 | -0.6456 | Yes | ||
| 45 | IFT172 | 3800372 7100400 | 17142 | -0.342 | -0.6248 | Yes | ||
| 46 | KIF3A | 5050348 6650079 | 17303 | -0.385 | -0.6073 | Yes | ||
| 47 | GNB1 | 2120397 | 17307 | -0.386 | -0.5812 | Yes | ||
| 48 | HDAC1 | 2850670 | 17368 | -0.409 | -0.5567 | Yes | ||
| 49 | GSK3B | 5360348 | 17444 | -0.435 | -0.5312 | Yes | ||
| 50 | PRKCD | 770592 | 17479 | -0.447 | -0.5026 | Yes | ||
| 51 | CSNK1E | 2850347 5050093 6110301 | 17665 | -0.518 | -0.4774 | Yes | ||
| 52 | LGALS3 | 2120239 | 17696 | -0.538 | -0.4425 | Yes | ||
| 53 | CREBBP | 5690035 7040050 | 17707 | -0.541 | -0.4063 | Yes | ||
| 54 | SIN3A | 2190121 | 18047 | -0.708 | -0.3765 | Yes | ||
| 55 | CSNK1G2 | 3060095 4730037 | 18124 | -0.763 | -0.3287 | Yes | ||
| 56 | PIAS1 | 5340301 | 18308 | -0.946 | -0.2743 | Yes | ||
| 57 | SPOP | 450035 | 18384 | -1.060 | -0.2063 | Yes | ||
| 58 | HDAC2 | 4050433 | 18463 | -1.273 | -0.1241 | Yes | ||
| 59 | SAP30 | 5080735 | 18567 | -1.947 | 0.0026 | Yes |